Introduction to the HLA System
If you’ve ever been interested in human immunology, then you’ve likely encountered the complicated and potentially confusing Human Leukocyte Antigen (HLA) system. Terms like “linkage disequilibrium” and “haplotype” may be enough to make your head spin!
You’re not alone if you’re not quite fluent in HLA-speak! To help, we’ve prepared a quick introduction to basic concepts and nomenclature of the HLA system. We hope you’ll find this to be a useful guide to better understand the HLA system and its role in immune defense and tissue matching.
Let’s start with the genetics of HLA. The genes which encode the HLA proteins are located within the Major Histocompatbility Complex (MHC), a genomic region on chromosome 6 which contains many important genes that function in the immune response. In fact, the MHC is the most genetically diverse region of the entire human genome.[1]
Located within the MHC, the HLA genes encode cell surface proteins that present peptide antigens to the immune system, thus enabling it to distinguish between self and foreign (e.g., pathogens).[2] So although you may hear someone use “HLA” and “MHC” interchangeably, you can look like an expert when you gently remind them that the MHC refers to the genomic region that contains many important immune function genes, including the HLA genes.
The MHC region is characterized by tight linkage (“disequilibrium”), which means that genes on the same chromosome tend to get passed to a person’s offspring together as a unit. This set of linked genes is often referred to as a “haplotype” and, due to the tight linkage of the MHC genes, we can often predict which HLA alleles a person might carry based on others that are typically found as part of that haplotype.
Classes of HLA Genes
The MHC region contains three classes of genes, and the HLA genes generally fall into two classes (I and II). The major classes of HLA genes and proteins are:
1. Class I HLA proteins: expressed on the cell surface of all nucleated cells in the human body. The role of class I HLA molecules is to present peptides derived from intracellular proteins.[3] In this way, class I HLA/peptide complexes enable immune surveillance on what is going on inside the cell, including viral infections and cancer. The most frequently studied (and genotyped) class I HLA genes are HLA-A, HLA-B, and HLA-C.
2. Class II HLA proteins: expressed by specialized cells of the immune system including dendritic cells, B cells, and macrophages (also known as antigen-presenting cells). The cells take up foreign antigens from the extracellular environment (bacteria, fungi, etc.) and digest them into small peptide fragments which can bind class II HLA proteins and be presented at the cell surface. T cells can recognize the class II HLA/peptide complexes, become activated, and secrete effector molecules (cytokines) which stimulate the adaptive immune response to the pathogen.[4] The most frequently studied (and genotyped) Class II HLA genes are HLA-DRB1/3/4/5, HLA-DQB1, HLA-DQA1, HLA-DPA1, and HLA-DPB1.
HLA Diversity: A Challenge for Nomenclature
The HLA genes are the among the most genetically diverse in the human genome. In fact, that is why they are so difficult to analyze using molecular techniques like DNA sequencing (more on this later).
Given the incredible diversity of microorganisms, perhaps it is no surprise that humans have evolved a polymorphic genetic system that can recognize these potential pathogens. However, immunity is a double-edged sword. The same diversity in the HLA system can also be a risk factor for the development of autoimmune diseases like type I diabetes. HLA differences between donors and recipients are also powerful mediators of the immune response to foreign tissues such as organ or bone marrow grafts.
To catalog the incredible diversity in the HLA system, a unique nomenclature system has been developed (Figure A) to describe each HLA allele that is present in the international database (IMGT/HLA) of all the known HLA alleles. At first glance, the nomenclature looks complicated, but it is pretty easy to understand once you know how it is structured:
Figure A. Human Leukocyte Antigen Nomenclature.
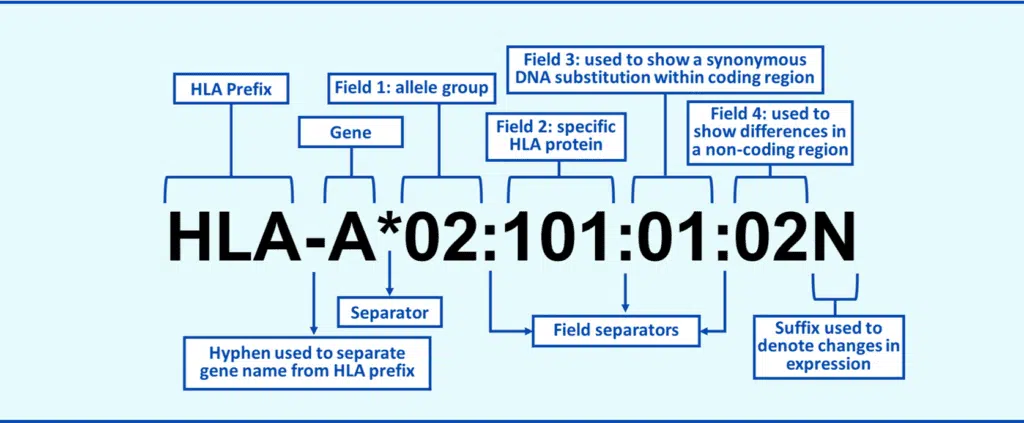
The “HLA” prefix and gene name (“A”, “B”, or “DRB1” for example) are self-explanatory, and the asterisk indicates you are looking at a molecular-level typing result. The rest of nomenclature is broken into “fields,” each giving you a different piece of information about the HLA allele. The first field equates to the antigen “family” of the allele. There are only so many HLA protein types (or antigens) and the first field basically tells you what antigen family the allele belongs to. For example, all A*01 alleles belong the A1 antigen family. Typing that gives you the first field is also called “antigen-level” or “low resolution” typing.
The second field tells captures non-synonymous DNA sequence changes that result in an amino acid difference in the protein. For example, A*02:01 differs from A*02:02 by at least one amino acid. Many people incorrectly assume that closely numbered second-field alleles are more similar. This is incorrect, as allele numbering relates to order in the database (date of allele discovery) not genetic similarity. For example, A*02:01 differs by three amino acids from A*02:02, not one. Typing that gives you the first and second fields is called “allele-level” or “high resolution” typing and is currently the standard used for bone marrow transplant matching.
The third and fourth fields provide additional information about the allele at the genetic level including synonymous changes that do not affect that amino acid sequence, and nucleotide changes that occur in untranslated regions of the gene (introns, etc.). Finally, information about the mature protein is captured as a suffix at the end of the allele nomenclature. Null (“N”) alleles are particularly important to identify as it indicates that the HLA protein is predicted not to be expressed at the cell surface.
HLA Nomenclature and Immunology Research
Understanding the basic genetics of the HLA system and how to describe and interpret HLA nomenclature is the first step in being able to use this information to enhance your immunology research. At Cytologics, we know how difficult learning the language of HLA can be. In future posts, we’ll share more insights as to relevance of HLA to your research including:
>> HLA typing: how is it done?
>> High resolution vs. low resolution typing: what do you really need for your experiment?
>> Role of HLA in cell therapy and vaccine research
>> Other essential knowledge about the HLA system
Conclusion
Basic and translational research across a broad spectrum of scientific fields frequently requires HLA-typed cells and other biospecimens. At Cytologics, we offer HLA typing information as part of the donor profile listed on the product certificate of analysis. We understand that HLA is vital to the outcomes of your experiments and we can help identify samples which meet your needs to include specific HLA types. Contact us today to discuss how we can support your research with HLA-typed immune cell products.
References
[1] Janeway CA Jr, Travers P, Walport M, et al. Immunobiology: The Immune System in Health and Disease. 5th edition. New York: Garland Science; 2001. The major histocompatibility complex and its functions. Available from: https://www.ncbi.nlm.nih.gov/books/NBK27156/.
[2] Apanius V, Penn D, Slev PR, Ruff LR, Potts WK. The nature of selection on the major histocompatibility complex. Crit Rev Immunol. 1997;17(2):179-224. doi: 10.1615/critrevimmunol.v17.i2.40. PMID: 9094452.
[3] Ibid.
[4] van Lith M, McEwen-Smith RM, Benham AM. HLA-DP, HLA-DQ, and HLA-DR have different requirements for invariant chain and HLA-DM. J Biol Chem. 2010;285(52):40800-40808. doi:10.1074/jbc.M110.148155.